
 |
|
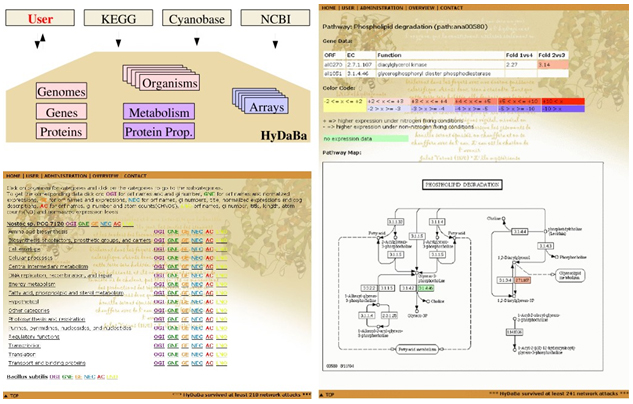
Figure 5. HyDaBa database. All experimental gene-expression data are saved in a relational database (http://www.hydaba.uni-koeln.de). This database allows cross-linking of the gene-expression data with genome data from NCBI and Cyanobase and metabolic charts available from KEGG. HyDaBa is based on a Apache Webserver, a MySQL database and a front-end programmed in PHP. All data are publicly accessible via this web interface. Upper left: database outline; Lower left: screen shot of metabolic categories; Right: screen shot of expression data overlayed on metabolic charts. |
|
|