|
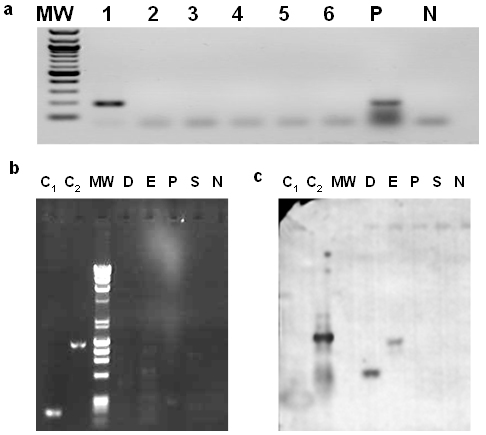
Figure 1. Identification of an
individual GM cotton seedling using PCR and Southern hybridization.
(a) Identification of an individual GM cotton seedling. PCR was performed
in each of the 6 plants (lanes 1-6) of a group tested positive for presence of
the CaMV 35S promoter. Lane 1 shows the band from the sample containing the 35S
sequence (195bp) similar to the positive control (P). MW is molecular weight
DNA marker and N is negative (no template) control.
(b) PCR products using the primers 35Spro3 and AP1 on the libraries
constructed with the Genome Walker kit (BD Biosciences, Palo Alto, USA) from
DNA of the GM seedling using the restriction enzymes DraI, EcoRV, PvuII and StuI (lanes D, E, P, S respectively). No clear bands
could be detected from the four libraries and the gel was transferred and
hybridized with a CaMV 35S promoter -specific probe. C1 and C2 are positive PCR controls using GM cotton genomic DNA as template and the
primers 35Spro5/35Spro3 and 35Spro5/NOS3 respectively and N is negative (no
template) control. MW is the λHind3-φx174 molecular weight DNA
marker.
(c) Southern hybridisation of a membrane produced after transfer of the PCR
products shown on the gel of Figure 1b with the CaMV 35S promoter -specific
probe. Lanes are as in Figure 1b. Two bands were detected corresponding to the
libraries D and E. Positive control C2 gave a strong hybridization
signal and C1 was overexposed and produced a blurred diffused band. |