|
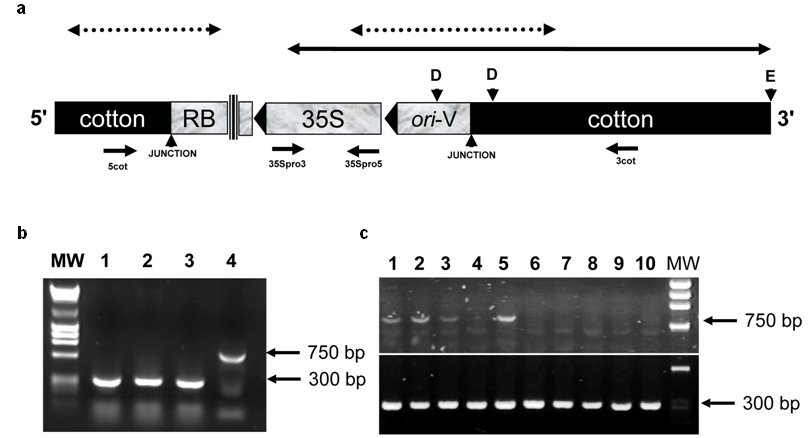
Figure 2. Genetic map and event-specific
PCR detection of transgenic cotton event 1445.
(a) Genetic map of part of the transgene construct and the flanking cotton
DNA fragments of the GM plant. The ori-V fragment and the CaMV 35S
promoter region of the construct are shown and arrowheads indicate the
direction towards the 3’ end of the elements. Vertical lines indicate
discontinuous map drawing. The solid double arrow above the map indicates the
fragment identified in this study while the dotted double arrows indicate
sequences reported in the Monsanto patent regarding cotton event
PV-GHGT07(1445) and compositions and methods for detection thereof (patent no
6,740,488, US Patent Office). D and E represent the restriction enzyme sites DraI,
and EcoRV, respectively. Junction sites are indicated by arrowheads
below the map of the construct and positions of primers are indicated with
arrows pointing towards the 3’ end of each primer.
(b) Event-specific PCR detection of cotton event 1445. Agarose gel analysis
of PCR products with the primers 3cot and 5cot, using as template three
different varieties of conventional cotton (lanes 1, 2, 3) and with the primers
35Spro3 and 3 cot and template transgenic event 1445 cotton (lane 4). MW is
molecular weight marker.
(c) Event-specific PCR detection of cotton event 1445 in seed samples tested positive for presence of the CaMV 35S promoter. Upper panel: PCR products
of a reaction performed with the 35Spro3 and 3cot primer pair that could
amplify the event 1445 specific 750 bp band in samples 1, 2, 3 and 5. In lane 1 the sample that was used as starting material of the study was loaded. Lower panel: PCR
products of a reaction performed with the 5cot and 3cot primer pair that could
amplify a cotton specific 300 bp band in all samples. MW is molecular weight
marker. |