
 |
|
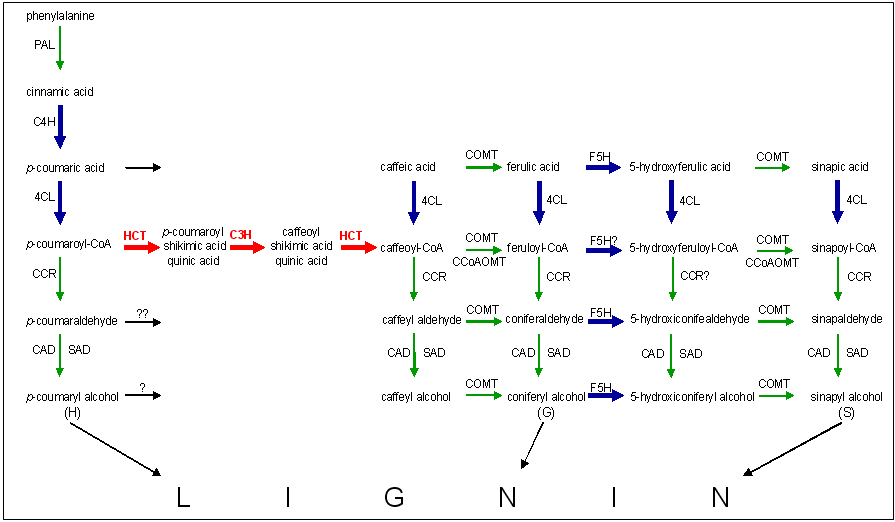
Figure 2. Reduction of total sulfide in effluent from a Figure 2. Simplified scheme of the enzymes proposed to be involved in lignin biosynthesis. Green arrows represent predicted E. globulus enzyme mediated reactions whose sequences are available in GenBank. Blue arrows represent sequences from other Eucalyptus species available in GenBank. Red arrows represent reactions catalyzed by enzymes that had not been previously described for any Eucalyptus species. Bold arrows correspond to sequences found in our E. globulus library that were not previously described for this species. Accesion numbers for E. globulus enzymes. PAL: phenylalanine ammonia lyase [GenBank: AF167487]; CCR: cinnamoyl CoA reductase [GenBank: DQ084797-DQ084795, GenBank: AH0154889]; CAD: cinnamyl alcohol dehydrogenase [GenBank: AF109157, AF038561 and O64969], COMT: caffeic acid O-methyltransferase [GenBank: Q9SWC2] and CCoAOMT: caffeoyl-CoA 3-O-methyltransferase [GenBank: AAD50443 and AAC26191). Accesion numbers for Eucalyptus enzymes, C4H: trans-cinnamate 4-hydroxylase [GenBank: AF149713-AF149715], F5H: ferulate 5-hydroxylase [GenBank: AJ249093; AF149713-AF149715] and 4CL: 4-coumarate: CoA ligase [GenBank: NC_003070, DQ147001, AJ244010, AF149715, AF149714 and AF149713]. reduction reactor by sulfur oxidizing bacteria. |
|
|