
 |
|
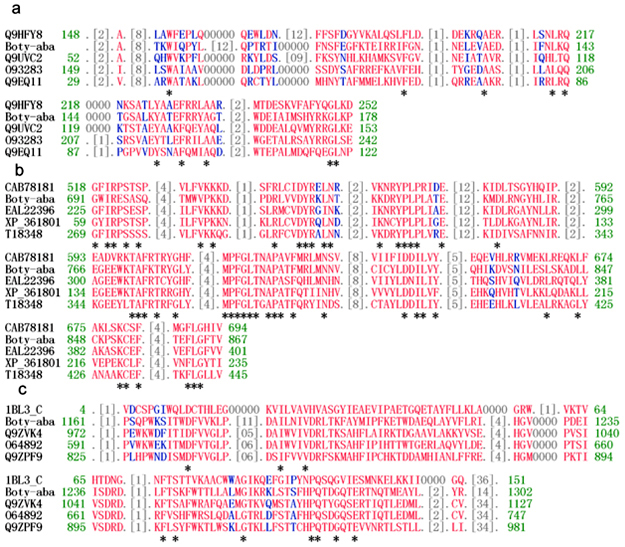
Figure
2. Analysis of the: Absolutely conserved amino acids are indicated with an asterisk (*), identical residues are coloured red, similar residues are coloured blue, masked out regions are coloured gray. Q9HFY8 is the gag protein of an LTR-retrotransposon (Cgret) from Glomerella cingulata; Q9UVC2 is the gag polyprotein of an LTR-retrotransposon (CfT-I) in Cladosporium fulvum; O93283 is the gag polyprotein of the LTR retrotransposon from Takifugu rubripes; Q9EQ11 is the gag domain of Myelin expression factor-3-like protein from Mus musculus; CAB78181 is the putative reverse-transcriptase-like protein from Arabidopsis thaliana; EAL22396 is the RT domain of the hypothetical protein CNBB2750 in Cryptococcus neoformans var. neoformans B-3501A; XP_361801 is the RT domain of the hypothetical protein MG04275.4 in Magnaporthe grisea 70-15; T18348 the RT domain of the gypsy retrotransposon in Magnaporthe grisea; 1BL3_C is the catalytic domain of HIV-1 integrase; Q9ZVK4 is the integrase core domain of the putative retroelement pol polyprotein from Arabidopsis thaliana; O64892 is the integrase core domain of the polyprotein from Ananas comosus; Q9ZPF9 is the IN domain of the F5K24.1 protein (putative polyprotein) from Arabidopsis thaliana. Retrotransposons are a widespread and important class of eukaryotic mobile genetic elements that have a central role in the structure, evolution, and function of eukaryotic genomes (Bennetzen, 2000; Kidwell and Lisch 2001). Recent reports have shown that retrotransposons contribute to the formation of genome structure and to the expression pattern of many host genes (Kashkush et al. 2003). For example, when Wis 2-1A retrotransposons are activated in wheat, the expression of several adjacent genes is activated or silenced by producing sense or antisense transcripts of those genes (Kashkush et al. 2003). Matsubara concluded that at least 3 transposable elements in Hf1 gene that plays a key role in the expression of floral colour in petunias govern anthocyanin biosynthesis of commercial petunias (Matsubara et al. 2005). It is not known whether Boty-aba has an affect on the expression of ABA genes. This divergence in structure of the ABA gene clusters gives new clues to survey the divergence in the ABA production yields of different B. cinerea strains. |
|
|