
 |
|
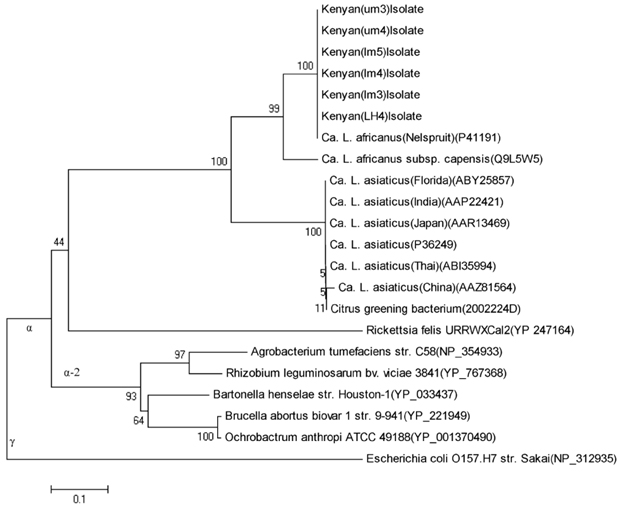
Figure 2. Minimum-evolution phylogenetic tree constructed from alignment of translated ribosomal protein L10 of ‘Candidatus Liberibacter’ species (accession numbers in parenthesis) and other αProteobacteria. The phylogenetic history was inferred using the Neighbor-Joining method. The evolutionary distances were computed using the Maximum Composite Likelihood method. Phylogenetic analyses were conducted in MEGA4. Bootstrap values are shown at the nodes. |
|
|