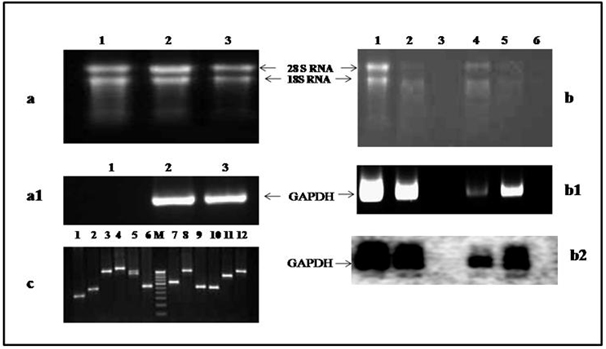
|
Figure 1. Qualitative and quantitative estimation of RNA isolated
by present and others methods.
(a) Total RNA isolated using present protocol from different
internodal tissues of Bambusa balcooa. RNA (approximately 1.0 µg each)
from 2nd; 5th and 10th in Lanes 1, 2 and 3
respectively were electrophorosed on 1.2% (w/v) formaldehyde denaturing agarose
gel. The 28S and 18S rRNA bands were indicated by arrows.
(a1) Electrophoresis of PCR amplified GAPDH gene on 1%
agarose gel using isolated RNA by present protocol as template (Lane 1); cDNA
prepared from isolated RNA as template (Lane 2) and genomic DNA of bamboo as
template (Lane 3).
(b) Comparison of total RNA isolated from 5th internode of Bambusa balcooa by different method on 1.2% (w/v)
formaldehyde denaturing agarose gel. Lanes 1, using present method; 2, using
method of Chan et al. 2007; 3, using method of Vasanthaiah et al. 2008; 4,
using method of Chiu et al. 2006; 5, by Trizol Reagent (Invitrogene, cat.#
10296010); 6, using method of Weng et al. 2009.
(b1) RT-PCR band (GADPH, 476 bp) obtained using RNA isolated
from 5th internode by different protocols. Lanes 1, using present
method; 2, using method of Chan et al. 2007; 3, using method of Vasanthaiah et al. 2008; 4, using method of Chiu et al. 2006; 5, from Trizol Reagent; and 6,
using method of Weng et al. 2009.
(b2) The GADPH gene was detected on northern blot using
total RNA isolated from 5th internodal bamboo tissue by different
methods. Lanes 1, using present method; 2, using method of Chan et al. 2007; 3,
using method of Vasanthaiah et al. 2008; 4, using method of Chiu et al. 2006;
5, by Trizol reagent; and 6, using method of Weng et al. 2009.
(c) A representation of subtraction cDNA library constructed
using RNA from 2nd and 10th internode of bamboo. PCR
amplifications of inserts obtained from different subtractive cDNA clones using
SP6 and T7 primers. PCR products were run with a 100 bp marker (Bangalore
Genei, cat.#105993) in a 1.0% agarose gel. |