
 |
|
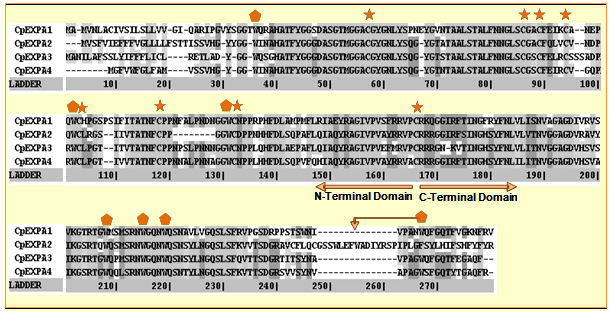
Fig. 3 Multiple sequence alignment of four homologs of CpEXPAs using Kalign and Kalignvu available at http://msa.cgb.ki.se/cgi-bin/msa.cgi. Ladder in the alignment is determined by the total number of aligned pairs divided by the maximum number of possible pairs. Shading over the amino acid sequence is based on the degree of conservation. First 165 amino acids comprise of N-terminal domain having signal peptide of varying length, while the remaining is shown as C-terminal domain. Eight conserved cystein (C) residues and seven tryptophan (W) residues are indicated by stars and pentagonals respectively. |
|
|