| | Molecular
Biology and Genetics |
| EJB Electronic Journal of Biotechnology ISSN:
0717-3458 | Vol.4 No. 3, Issue of December 15,
2001. | | © 2001 by Universidad Católica
de Valparaíso -- Chile | Received
July 12, 2001 / Accepted November 7, 2001 |
Molecular dynamics simulations
of active site mutants of rat liver arginase Mauricio
Canales*
Laboratorio de Biofísica,
Departamento de Biología Molecular
Facultad de Ciencias Biológicas,
Universidad de Concepción
Casilla 152-C Correo 3, Concepción,
Chile
Tel: 56 41 203822
Fax: 56 41 239687
E-mail: mcanale@udec.cl Linda
Westermeyer
Facultad
de Agronomía, Universidad Adventista de Chile
Casilla 7-D, Chillán,
Chile
Nelson
Carvajal
Laboratorio
de Enzimología, Departamento de Biología Molecular
Facultad
de Ciencias Biológicas, Universidad de Concepción
Casilla 160-C
Correo 3, Concepción, Chile
Tel: 56 41 203814
Fax: 56 41 968723
E-mail: ncarvaja@udec.cl *
Corresponding author
Financial
support: Project 4533 from Universidad de Concepción,
Chile to M.C.A.
Keywords: arginase, arginine, molecular
dynamics, optimization.
By
using molecular dynamics (MD) simulations and crystallographic data for rat liver
arginase, the substrate positions in the active sites of native and mutant forms
of the enzyme, were compared and correlated with known kinetic consequences of
mutations. The mutants compared were His 141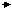 Phe
and His 141 Phe
and His 141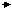 Asn.
The simulations show that mutation His141 Asn.
The simulations show that mutation His141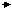 Asn
gives the greatest divergence from the atomic coordinates, when compared with
the control native enzyme. The mutant Asp128 Asn
gives the greatest divergence from the atomic coordinates, when compared with
the control native enzyme. The mutant Asp128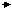 Asn
does not show a change in atomic coordinates in the substrate, in agreement with
the concept that a change in the metal coordination is responsible for the loss
of catalytic activity in this mutant. Results obtained agree with reported kinetic
consequences of mutations in arginase. Asn
does not show a change in atomic coordinates in the substrate, in agreement with
the concept that a change in the metal coordination is responsible for the loss
of catalytic activity in this mutant. Results obtained agree with reported kinetic
consequences of mutations in arginase.
|

